Our research aims at developing low-dose electron microscopy techniques to achieve high-resolution 3D imaging while minimizing radiation damage, particularly in biological materials.
We apply cryo-electron microscopy, STEM tomography, 4DSTEM and electron ptychography to study radiation sensitive materials, with a strong emphasis on bio-related applications. In addition, we are exploring methods for volume imaging of thick samples with minimal sample distortion to advance our understanding of complex cellular systems.
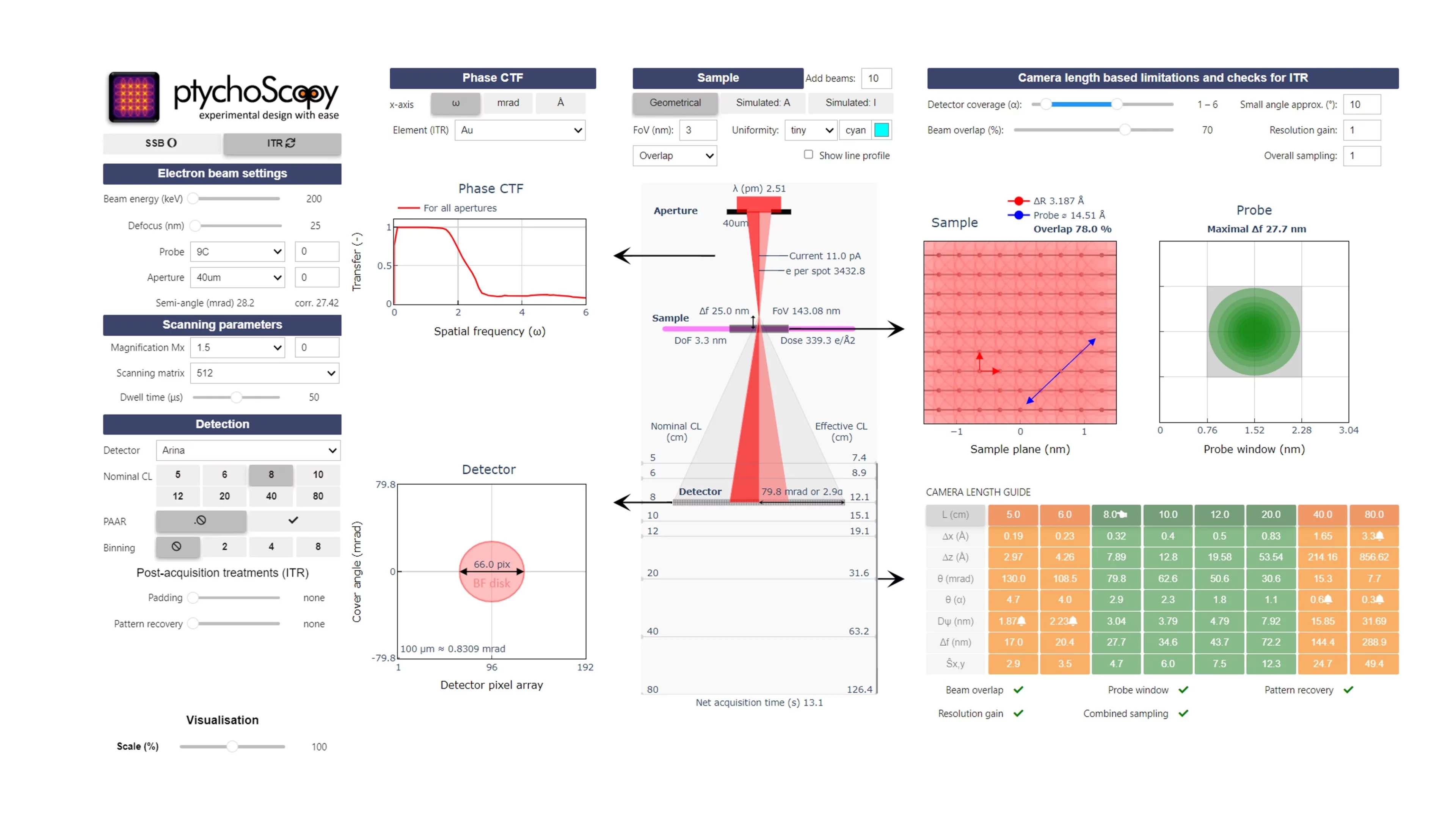
One example of our recent work is ptychoScopy, a Python-based software tool that we developed to support the setup and optimization of electron ptychography experiments. The tool helps researchers to navigate key experimental parameters—such as beam convergence, defocus, and dose—to improve reconstruction quality. By guiding users through the complex design process, ptychoScopy facilitates more efficient and successful ptychographic imaging across a range of scientific applications.
References:
-
Aidukas T, Phillips NW, Diaz A, Poghosyan E, Müller E, Levi AFJ, et al.
High-resolution, data-driven 3D X-ray imaging of microchips using ptychography
In: El Gawhary O, Witte S, Moreno I, eds. 2025 European Optical society annual meeting (EOSAM 2025). Vol. 335. EPJ Web of Conferences. Les Ulis Cedex A: EDP Sciences; 2025:01001 (2 pp.). https://doi.org/10.1051/epjconf/202533501001
DORA PSI -
Skoupy R, Müller E, Pennycook TJ, Guizar-Sicairos M, Fabbri E, Poghosyan E
Ptychoscopy: a user friendly experimental design tool for ptychography
Scientific Reports. 2025; 15(1): 24959 (15 pp.). https://doi.org/10.1038/s41598-025-09871-6
DORA PSI -
Aidukas T, Phillips NW, Diaz A, Poghosyan E, Müller E, Levi AFJ, et al.
High-performance 4-nm-resolution X-ray tomography using burst ptychography
Nature. 2024; 632(8023): 81-88. https://doi.org/10.1038/s41586-024-07615-6
DORA PSI -
Alvarez N, Bruggmann R, Buchmann N, Dessimoz C, Faso C, Hofmann S, et al.
Biology community roadmap 2024. Update of Swiss community needs for research infrastructures 2029-2032
Bern: Swiss Academy of Sciences (SCNAT); 2024. Swiss Academies reports: 19/6. https://doi.org/10.5281/zenodo.14264965
DORA PSI -
Skoupy R, Grando Stroppa D, Guizar-Sicairos M, Mueller E, Fabbri E, Poghosyan E
Newcomer's guide into optimal data acquisition for electron ptychography
In: Celio G, LeBeau J, Evans J, Spurgeon S, Guy N, eds. 2024 proceedings microcospy and microanalysis. Vol. 30. Microscopy and Microanalysis. Oxford University Press; 2024:1846-1847. https://doi.org/10.1093/mam/ozae044.913
DORA PSI -
Xie X, Barba Flores L, Bejar Haro B, Bergamaschi A, Fröjdh E, Müller E, et al.
Enhancing spatial resolution in MÖNCH for electron microscopy via deep learning
Journal of Instrumentation. 2024; 19(01): C01020 (11 pp.). https://doi.org/10.1088/1748-0221/19/01/C01020
DORA PSI -
Collu G, Bierig T, Krebs A-S, Engilberge S, Varma N, Guixà-González R, et al.
Chimeric single α-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology
Structure. 2022; 30(1): 95-106. https://doi.org/10.1016/j.str.2021.09.002
DORA PSI -
Cvjetinović Đ, Janković D, Milanović Z, Mirković M, Petrović J, Prijović Ž, et al.
177Lu–labeled micro liposomes as a potential radiosynoviorthesis therapeutic agent
International Journal of Pharmaceutics. 2021; 608: 121106 (10 pp.). https://doi.org/10.1016/j.ijpharm.2021.121106
DORA PSI -
Marelli E, Gazquez J, Poghosyan E, Müller E, Gawryluk DJ, Pomjakushina E, et al.
Correlation between oxygen vacancies and oxygen evolution reaction activity for a model electrode: PrBaCo2O5+δ
Angewandte Chemie International Edition. 2021; 60(26): 14609-14619. https://doi.org/10.1002/anie.202103151
DORA PSI -
Bierig T, Collu G, Blanc A, Poghosyan E, Benoit RM
Design, expression, purification, and characterization of a YFP-tagged 2019-n CoV spike receptor-binding domain construct
Frontiers in Bioengineering and Biotechnology. 2020; 8: 618615 (10 pp.). https://doi.org/10.3389/fbioe.2020.618615
DORA PSI -
Poghosyan E, Iacovache I, Faltova L, Leitner A, Yang P, Diener DR, et al.
The structure and symmetry of the radial spoke protein complex in Chlamydomonas flagella
Journal of Cell Science. 2020; 133(16): jcs245233 (9 pp.). https://doi.org/10.1242/jcs.245233
DORA PSI -
Zhu X, Poghosyan E, Rezabkova L, Mehall B, Sakakibara H, Hirono M, et al.
The roles of a flagellar HSP40 ensuring rhythmic beating
Molecular Biology of the Cell. 2019; 30(2): 228-241. https://doi.org/10.1091/mbc.E18-01-0047
DORA PSI -
Zhu X, Poghosyan E, Gopal R, Liu Y, Ciruelas KS, Maizy Y, et al.
General and specific promotion of flagellar assembly by a flagellar nucleoside diphosphate kinase
Molecular Biology of the Cell. 2017; 28(22): 3029-3042. https://doi.org/10.1091/mbc.E17-03-0156
DORA PSI
Current team members:
Former team members:
Evan Quistad (Bachelor student)
Dr. Radim Skoupy (Postdoc)
Contact
Lab. for Multiscale Bioimaging
Paul Scherrer Institute PSI
Forschungsstrasse 111
5232 Villigen PSI, Switzerland