Biography
Roger leads the Biomolecular Nanoengineering research team. He holds degrees in Molecular Biology and Biophysics from the University of Colorado Boulder, the University of Basel, and the University of Salzburg, with doctoral work conducted in collaboration with Novartis. His training spans structural biology, protein engineering, and biophysics, with early work in electron microscopy followed by extensive experience in crystallography and structure-based drug discovery at the Novartis Institutes for BioMedical Research. As a postdoctoral researcher at the Paul Scherrer Institute (PSI), he expanded his expertise to membrane proteins and developed a research focus on the structure based design and engineering of functional fusion proteins, including molecular biomimetics and de novo protein design. His current research centers on protein-based tools for multiscale bioimaging, including tags to localize proteins, enhance imaging contrast, and generate molecular barcodes, with particular emphasis on enzymes and receptors involved in blood pressure regulation and inflammation during ageing, such as the β1-adrenergic receptor and angiotensin-converting enzyme 2.
Research
New findings
- Through mRNA display, we identified a novel macrocyclic peptide that binds in the ACE2 catalytic cleft, distant from the catalytic zinc ion site, and locks it in an open conformation. The peptide, WJL-63, holds promise as a targeting agent for tracers for multiscale bioimaging and as a tool compound to study ACE2 conformation. A preprint is available on bioRxiv: https://www.biorxiv.org/content/10.64898/2025.12.01.690145v1
- By filtering the protein data bank for relatively large proteins with optimal geometry for genetic insertion into intracellular loop 3 of class A GPCRs, we identified AmpC β-lactamase as a top hit. We designed a fusion protein in which this protein is connected to β1 adrenergic receptor via two rigid chimeric helices. This addition to the receptor provides sufficient mass and discernable structural features to enable a good signal-to-noise ratio and precise particle alignment in cryo-EM. In the newest version of the pre-print, we improved the cryo-EM structure, allowing visualization of the bound small molecule compound: https://www.biorxiv.org/content/10.1101/2021.09.25.461805v2
Structure-function relationships of ACE2 across multiple biological scales
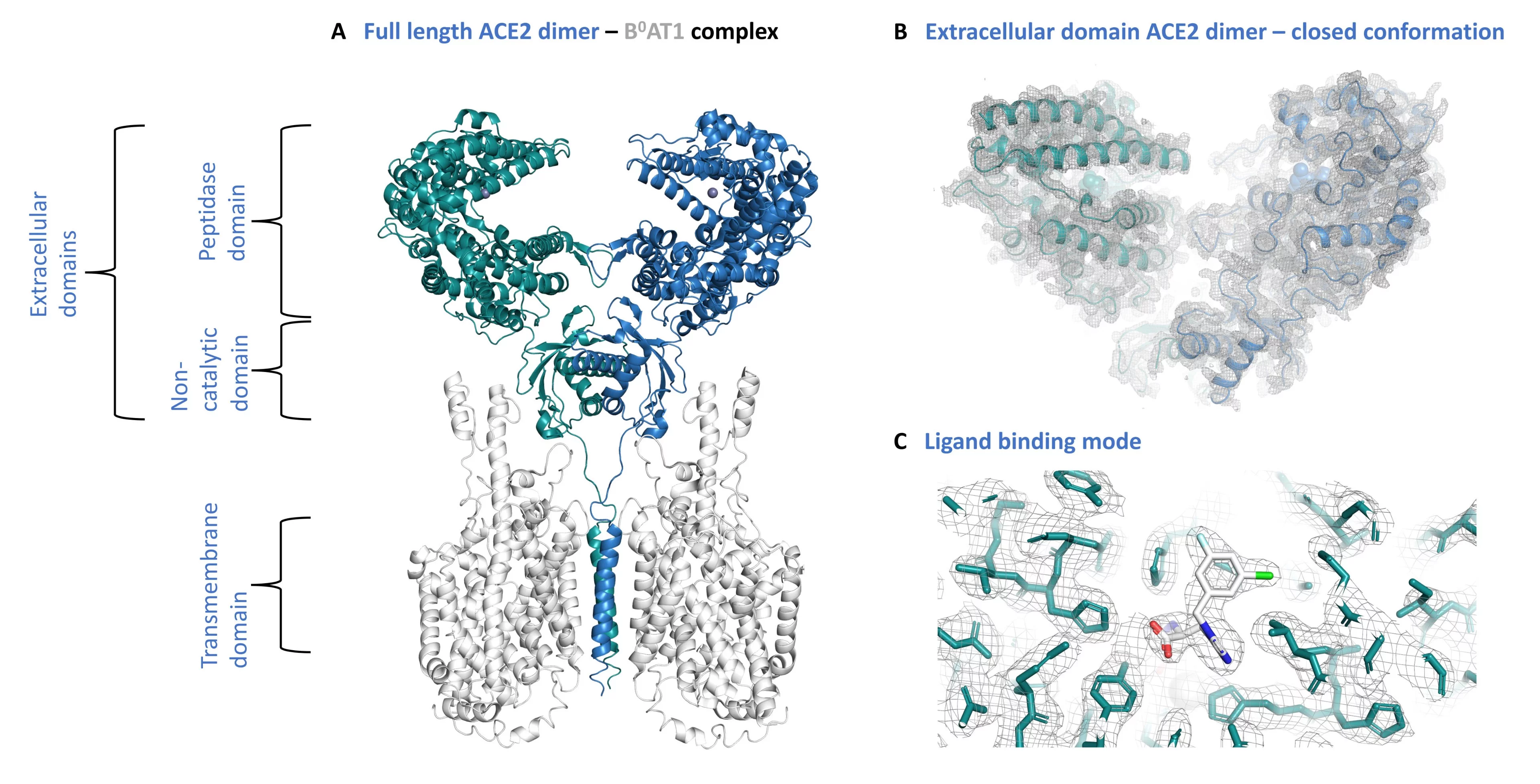
The protein angiotensin-converting enzyme 2 (ACE2), a zinc metallopeptidase, plays key roles in physiological processes involved in health and disease. It became widely known as the receptor used by SARS-CoV and SARS-CoV-2 for cell recognition and entry (Zhou et al., 2020, PMID: 32015507). ACE2 is a main node in the protective arm of the renin-angiotensin-aldosterone system (RAAS) of blood pressure control, and is involved in development, inflammation, immunity, and neurodegenerative deseases (PMID: 23686164). ACE-like proteins have also been identified as components in venoms, for example from scorpions or spiders (PMID: 331463). In addition to its peptidase activities, ACE2 is involved in other processes, such as the regulation of interstinal neutral amino acid transport (PMID: 21814048).
The enzyme is gaining increased attention as a drug target for hypertension, cardiovascular disease, diabetes, and other disorders. ACE2 is present on the cell surface of various tissues with a specific distribution and can be released into solution by sheddases (PMID: 15983030).
Structurally, the extracellular or soluble part of ACE2 comprises an N-terminal peptidase domain with a wide, deep substate binding cleft flanking the conserved Zinc-ion binding site, followed by a smaller non-catalytic domain (PMID: 14754895). In the full-length protein, the non-catalytic domain domain extends into a single transmembrane helix and an intracellular domain.
My research focuses on the study of the structure, conformation, and biophysics of ACE2 in the context of novel peptides and small molecule inhibitors, and its interactions with the receptor-binding domain (RBD) of the SARS-CoV-2 spike (S) glycoprotein. Another important aspect towards a detailed understanding of ACE2 function is its ultrastructure and interactions in a cellular context across different scales, ranging from molecules to cells and tissues. This requires identification of the protein of interest in its natural, crowded environment of cells.
Beamlines and tools
Exploring the function of ACE2 (or other proteins) across multiple scales requires access to a wide range of specialized equipment, and interdisciplinary collaborations.
Our department is equipped with state-of-the-art light- and electron microscopy facilities, allowing the tracking of proteins in cells. Access to various beamlines is of key importance. The PSI macromolecular crystallography beamlines X06SA (PXI) and X06DA (PXIII) provide bright collimated X-ray beams to study protein structure and interactions with ligands at near-atomic resolution. The beamline X02DA (TOMCAT) has been constructed for X-ray imaging by synchrotron tomographic microscopy, which enables X-ray imaging of larger objects such as whole cells or tissues. The SwissFEL X-ray free electron laser (SwissFEL) generates a very intense and focused beam of X-rays at short pulses, enabling the study time-resolved changes in protein structures.
Team members
Education
Biochemie und Molekularbiologie - SpringerSpektrum - 2024, 2nd Edition
Christen - Jaussi - Benoit
Publications
-
Benoit RM
Editorial: fusion proteins for the detection of pathogens or pathogen receptors
Frontiers in Bioengineering and Biotechnology. 2025; 13: 1660729 (3 pp.). https://doi.org/10.3389/fbioe.2025.1660729
DORA PSI -
Gupta R, Schärer P, Liao Y, Roy B, Benoit RM, Shivashankar GV
Regulation of p65 nuclear localization and chromatin states by compressive force
Molecular Biology of the Cell. 2025; 36(4): 36:ar37 (12 pp.). https://doi.org/10.1091/mbc.E23-11-0431
DORA PSI -
Christen P, Jaussi R, Benoit R
Biochemie und Molekularbiologie. Eine Einführung in 40 Lerneinheiten
2nd ed. Berlin: Springer Nature; 2024. https://doi.org/10.1007/978-3-662-65477-4
DORA PSI -
Wang J, Beyer D, Vaccarin C, He Y, Tanriver M, Benoit R, et al.
Development of radiofluorinated MLN-4760 derivatives for PET imaging of the SARS-CoV-2 entry receptor ACE2
European Journal of Nuclear Medicine and Molecular Imaging. 2024; 52: 9-21. https://doi.org/10.1007/s00259-024-06831-6
DORA PSI -
Behbahanipour M, Benoit R, Navarro S, Ventura S
OligoBinders: bioengineered soluble amyloid-like nanoparticles to bind and neutralize SARS-CoV-2
ACS Applied Materials and Interfaces. 2023; 15(9): 11444-11457. https://doi.org/10.1021/acsami.2c18305
DORA PSI -
Farnung J, Muhar M, Liang JR, Tolmachova KA, Benoit RM, Corn JE, et al.
Semisynthetic LC3 probes for autophagy pathways reveal a noncanonical LC3 interacting region motif crucial for the enzymatic activity of human ATG3
ACS Central Science. 2023; 9(5): 1025-1034. https://doi.org/10.1021/acscentsci.3c00009
DORA PSI -
Collu G, Bierig T, Krebs A-S, Engilberge S, Varma N, Guixà-González R, et al.
Chimeric single α-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology
Structure. 2022; 30(1): 95-106. https://doi.org/10.1016/j.str.2021.09.002
DORA PSI -
Bierig T, Collu G, Blanc A, Poghosyan E, Benoit RM
Design, expression, purification, and characterization of a YFP-tagged 2019-n CoV spike receptor-binding domain construct
Frontiers in Bioengineering and Biotechnology. 2020; 8: 618615 (10 pp.). https://doi.org/10.3389/fbioe.2020.618615
DORA PSI -
Skopintsev P, Ehrenberg D, Weinert T, James D, Kar RK, Johnson PJM, et al.
Femtosecond-to-millisecond structural changes in a light-driven sodium pump
Nature. 2020; 583: 314-318. https://doi.org/10.1038/s41586-020-2307-8
DORA PSI -
Krebs A-S, Bierig T, Collu G, Benoit RM
Seamless insert-plasmid assembly at sub-terminal homologous sequences
Plasmid. 2019; 106: 102445 (9 pp.). https://doi.org/10.1016/j.plasmid.2019.102445
DORA PSI -
Benoit RM
Botulinum neurotoxin diversity from a gene-centered view
Toxins. 2018; 10(8): 310 (14 pp.). https://doi.org/10.3390/toxins10080310
DORA PSI -
Benoit RM, Schärer MA, Wieser MM, Li X, Frey D, Kammerer RA
Crystal structure of the BoNT/A2 receptor-binding domain in complex with the luminal domain of its neuronal receptor SV2C
Scientific Reports. 2017; 7: 43588 (7 pp.). https://doi.org/10.1038/srep43588
DORA PSI -
Heydenreich FM, Miljuš T, Jaussi R, Benoit R, Milić D, Veprintsev DB
High-throughput mutagenesis using a two-fragment PCR approach
Scientific Reports. 2017; 7: 6787 (11 pp.). https://doi.org/10.1038/s41598-017-07010-4
DORA PSI -
Benoit RM, Ostermeier C, Geiser M, Li JSZ, Widmer H, Auer M
Seamless insert-plasmid assembly at high efficiency and low cost
PLoS One. 2016; 11(4): e0153158 (13 pp.). https://doi.org/10.1371/journal.pone.0153158
DORA PSI -
Bianchi S, van Riel WE, Kraatz SHW, Olieric N, Frey D, Katrukha EA, et al.
Structural basis for misregulation of kinesin KIF21A autoinhibition by CFEOM1 disease mutations
Scientific Reports. 2016; 6: 30668 (16 pp.). https://doi.org/10.1038/srep30668
DORA PSI -
Christen P, Jaussi R, Benoit R
Biochemie und Molekularbiologie. Eine Einführung in 40 Lerneinheiten
Berlin, Heidelberg: Springer; 2016. https://doi.org/10.1007/978-3-662-46430-4
DORA PSI -
Benoit RM, Frey D, Wieser MM, Thieltges KM, Jaussi R, Capitani G, et al.
Structure of the BoNT/A1 - Receptor complex
Toxicon. 2015; 107(Part A): 25-31. https://doi.org/10.1016/j.toxicon.2015.08.002
DORA PSI -
Benoit RM, Frey D, Hilbert M, Kevenaar JT, Wieser MM, Stirnimann CU, et al.
Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A
Nature. 2014; 505(7481): 108-111. https://doi.org/10.1038/nature12732
DORA PSI -
Kammerer RA, Benoit RM
Botulinum neurotoxins: new questions arising from structural biology
Trends in Biochemical Sciences. 2014; 39(11): 517-526. https://doi.org/10.1016/j.tibs.2014.08.009
DORA PSI -
Benoit RM, Meisner N-C, Kallen J, Graff P, Hemmig R, Cèbe R, et al.
The X-ray crystal structure of the first RNA recognition motif and site-directed mutagenesis suggest a possible hur redox sensing mechanism
Journal of Molecular Biology. 2010; 397(5): 1231-1244. https://doi.org/10.1016/j.jmb.2010.02.043
DORA PSI
Benoit RM and Auer M
A direct way of redox sensing
RNA Biol 2011;8:18-23
DOI: 10.4161/rna.8.1.13555
Meisner N.C., Hintersteiner M., Seifert J.M., Bauer R., Benoit R.M., Widmer A., Schindler T., Uhl V., Lang M., Gstach H., Auer M.
Terminal adenosyl transferase activity of posttranscriptional regulator HuR revealed by confocal on-bead screening.
J Mol Biol. 2009; 386: 435-450
DOI: 10.1016/j.jmb.2008.12.020
Benoit, R.M., Wilhelm, R.N., Scherer-Becker, D., Ostermeier, C.
An improved method for fast, robust, and seamless integration of DNA fragments into multiple plasmids.
Protein Expr Purif. 2006; 45: 66-71
DOI: 10.1016/j.pep.2005.09.022