Group website: Cellular structural biology and imaging
Research by Takashi Ishikawa
We apply these methods to reveal the mechanism of flagellar/ciliary bending motion. In flagella/cilia, which consist of ~300 proteins, nine microtubule doublets are linked by dynein motor proteins. We are analyzing molecular arrangement of dynein and other proteins by electron tomography and refine high resolution structures by single particle analysis, developing methodologies.
Eukaryotic flagella/cilia are bending organelles which drive cells forward or backward, or generate extracellular flows. They exist in sperms, tracheae (respiratory cilia), brain, embryo (nodal cilia) and many other organs. Therefore defects of flagella/cilia cause diseases (ciliopathy). Flagella/cilia are attractive in the view point of nanomachine development.
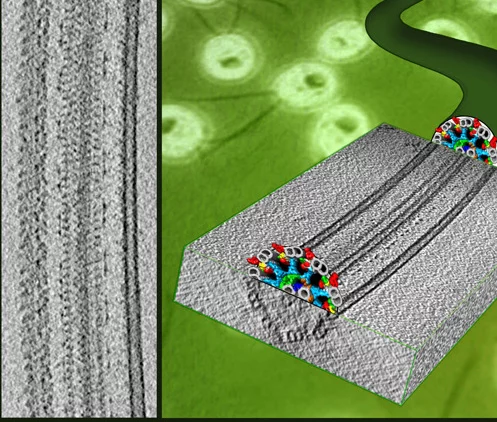
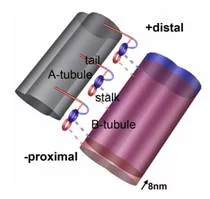
Flagella and cilia from various species and organisms share the “9+2” axonemal structure, in which nine microtubule doublets surround two singlet microtubules. Adjacent microtubule doublets slide past each other by ATP-driven power stroke of dynein motor proteins. Our research focuses the mechanism of force generation of dynein and the mechanism to integrate dynein power strokes into well-orchestrated bending motion of flagella/cilia. We employed technique of electron cryo-tomography (fig. 1) and 3D image analysis to answer these questions.
3D molecular architecture of outer and inner dynein arms from Chlamydomonas flagella
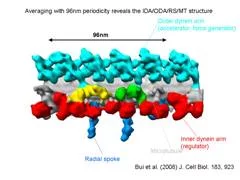
We extracted 96nm periodic fragments (subtomograms) from tomograms of flagella (fig. 1), aligned and averaged them three-dimensionally (fig. 2). The obtained 3D structure describes the molecular conformation and arrangement of dyneins and other molecules on the microtubule doublets. In the inner dynein arm (which determines the wave form) eight dyneins form a longitudinal array, while the outer arm (responsible for force generation and acceleration) consists of three dyneins stacking vertically. At ~35Å resolution, our averaged tomograms revealed 3D conformation of dynein (which consists of an AAA-ring (ring-shaped ATPase domain), a coiled-coil stalk and an N-terminal tail) in situ. N-terminal tails extend from AAA-rings toward the distal end (= plus end of the microtubule). By combining 3D structures of mutants which lack various dynein isoforms, we identified them in our 3D map (fig. 2).
Bui, K.H., Sakakibara, H., Movassagh, T., Oiwa, K. and Ishikawa, T. (2008) “Molecular architecture of inner dynein arms in situ in Chlamydomonas flagella” J. Cell Biol., 183, 923-932. Ishikawa, T., Sakakibara, H., and Oiwa, K. (2007) “The architecture of outer dynein arms in situ” J. Mol. Biol. 368, 1249-1258.
Structural basis of dynein power stroke and flagellar/ciliary bending
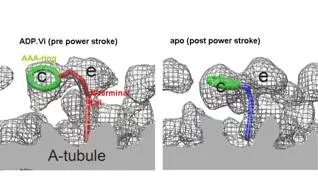
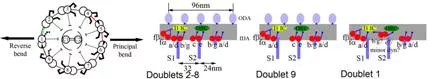
Averaged tomograms of flagella in the presence and in the absence of nucleotides showed the conformational change of dyneins. In the presence of ADP.Vi (Vi: vanadate), which is non-hydrolysable ATP analogue and mimics the pre-power stroke state, the AAA-ring of dynein stays at the proximal (minus end of the microtubule) and the N-terminal tail gradually curves toward the microtubule. However, in the apo state (corresponding to the post-power stroke state), the ATPase head moves toward the distal (plus) end and the N-terminal tail sharply kinks toward the microtubule. This indicates the translational shift of dynein (not rotation), pulls the adjacent microtubule toward the distal end (fig. 3).
In the flagella in the presence of ADP.Vi, this conformational change occurs at about half of dyneins, while the other dyneins stay at the apo conformation, indicating cooperativity among dyneins prohibits other dyneins to change their conformations. Interestingly these two conformations do not appear randomly, but form cluster (fig. 4). This phenomenon provides one hypothesis of flagellar/ciliary bending: torsion is generated at interface between two clusters.
Movassagh, T., Bui, K.H., Sakakibara, H., Oiwa, K. and Ishikawa, T. (2010) “Global conformational changes of dynein arms in flagella induced by nucleotides” Nat. Struct. Mol. Biol., 17, 761-767.
Asymmetry of dynein arrangement in Chlamydomonas flagella – mechanism of planar asymmetric bending
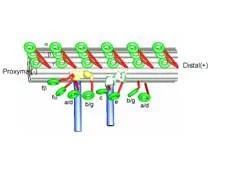
We found that the arrangement of inner arm dyneins is not perfectly symmetrical among nine microtubule doublets of the Chlamydomonas flagellum. Two microtubule doublets, which are apposed between two flagella of one Chlamydomonas cell, lacks a few inner arm dyneins (fig. 5), which will results in asymmetric force generation between the internal and external sides of flagellar bending. Our tomographic studies identified linkers which connect only doublets on the bending plane, suggesting the mechanism to limit the flagellar motion planar.
Bui, K.H., Sakakibara, H., Movassagh, T., Oiwa, K. and Ishikawa, T. (2009) “Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella” J. Cell Biol. 186, 437-446.
Our main projects
Bui, K.H., Toshiki, Y., Yamamoto, R., Kamiya, R. and Ishikawa, T. (2012) “Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme.” J. Cell Biol., 198, 913-925.
Maheshwari, A. and Ishikawa, T. (2012) “Heterogeneity of dynein structure implies coordinated suppression of dynein motor activity in the axoneme.” J. Struct. Biol., 179, 235-241.
Ueno, H., Ishikawa, T., Bui, K.H., Gonda, K., Ishikawa, T. and Yamaguchi, T. (2012) “Mouse respiratory cilia with the asymmetric axonemal structure on sparsely distributed ciliary cells can generate overall directional flow.” Nanomedicine, 8, 1081-1087.
Pigino, G., Maheshwari, A., Bui, K.H., Shingyoji, C., Kamimura, S. and Ishikawa, T. (2012) “Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena and sea urchins.” J. Struct. Biol., 178, 199-206.
Guichard, P., Desfosses, A., Maheshwari, A., Hachet, V., Dietrich, C., Brune, A., Ishikawa, T., Sachse, C. and Gonczy, P. (2012) “Cartwheel architecture of Trichonympha basal body.” Science, 337, 553.
Pigino, G., Bui, K.H., Maheshwari, A., Lupetti, P., Diener, D. and Ishikawa, T. (2011) “Cryoelectron tomography of radial spokes in cilia and flagella.” J. Cell Biol., 195, 673-687.
Bui, K.H., Pigino, G. and Ishikawa, T. (2011) “3D structural analysis of eukaryotic flagella/cilia by electron cryo-tomography” J. Synchrotron Radiation, 18, 2-5.
Movassagh, T., Bui, K.H., Sakakibara, H., Oiwa, K. and Ishikawa, T. (2010) “Global conformational changes of dynein arms in flagella induced by nucleotides” Nat. Struct. Mol. Biol., 17, 761-767.
Bui, K.H., Sakakibara, H., Movassagh, T., Oiwa, K. and Ishikawa, T. (2009) “Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella” J. Cell Biol. 186, 437-446.
Bui, K.H., Sakakibara, H., Movassagh, T., Oiwa, K. and Ishikawa, T. (2008) “Molecular architecture of inner dynein arms in situ in Chlamydomonas flagella” J. Cell Biol., 183, 923-932.
Ishikawa, T., Sakakibara, H., and Oiwa, K. (2007) “The architecture of outer dynein arms in situ” J. Mol. Biol. 368, 1249-1258.
Reviews and book chapters
Ishikawa, T. (2012) “Structural biology of cytoplasmic and axonemal dyneins.“ J. Struct. Biol. 179, 229-234.
Pigino, G. and Ishikawa, T. (2012) “Axonemal radial spokes: 3D structure, function and assembly“ BioArchitecture, 2, 50-58.
Ishikawa, T. (2012) “3D structures of axonemes” in “Handbook of Dynein” edited by L. Amos and K. Hirose, Pan Stanford
Ishikawa, T. (2011) “Organization of dyneins and associated regulatory systems in the axoneme” in “Dyneins: Structure, biology and disease” edited by S. King, Elsevier
Collaborations with other groups
Liebi, M., Kuster, S., Kohlbrecher, J., Ishikawa, T., Fischer, P., Walde, P. and Windhab, E.L. (2012) “Magnetically enhanced bicelles delivering switchable anisotropy in optical gels.“ ACS Appl. Mater Interfaces, 6, 1100-1105.
Liebi, M., Kohlbrecher, J., Ishikawa, T., Fischer, P., Walde, P. and Windhab, E.L. (2012) “Cholesterol increases the magnetic aligning of bicellar disks from an aqueous mixture of DMPC and DMPE-DTPA with complexed thulium ions.“ Langmuir, 28, 10905-10915.
Lowell, A.N., Qiao, H., Liu, T., Ishikawa, T., Zhang, H., Oriana, S., Wang, M., Ricciotti, E., FitzGerald, G.A., Zhou, R. and Yamakoshi, Y. (2012) “Functionalized low-density lipoprotein nanoparticles for in vivo enhancement of atherosclerosis on magnetic resonance images“ Bioconjug. Chem., 23, 2313-2319.
Tokutsu, R., Kato, N., Bui, K.H., Ishikawa, T. and Minagawa, J. (2012) “Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii“ J. Biol. Chem., 287, 31574-31581.
Effantin, G., Ishikawa, T., De Donatis, G.M., Maurizi, M.R. and Steven, A.C. (2010) “Local and global mobility in th ClpA AAA+ chaperone detected by cryo-electron microscopy: functional connotations“ Structure, 18, 553-562.
Megli, P., Conte, E. and Ishikawa, T. (2010) “Cholesterol attenuates and prevents bilayer damage and breakdown in lipoperoxidized model membranes. A spin labeling EPR study“ Biochim. Biophys. Acta, 1808, 2267-2274.
Beck, P., Liebi, M., Kohlbrecher, J., Ishikawa, T., Ruegger, H., Zepik, H, Fischer, P., Walde, P. and Windhab, E. (2010) “Novel type of bicellar disks from a mixture of DMPC and DMPE-DTPA with complexed lanthanides“ Langmuir, 26, 5382-5387.
Bleicken, S., Classen, M., Padmavathi, P.V., Ishikawa, T., Zeth, K., Steinhoff, H.J. and Bordignon, E. (2010) “Molecular details of Bax activation, oligomerization and membrane insertion“ J. Biol. Chem. 285, 6636-6647.
Beck, P., Liebi, M., Kohlbrecher, J., Ishikawa, T., Ruegger, H., Zepik, H, Fischer, P., Walde, P. and Windhab, E. (2010) “Magnetic field alignable domains in phospholipid vesicle membranes containing lanthanides“ J. Phys. Chem. B 114, 174-186.
Guo, Z.W., Ruegger, H., Kissner, R., Ishikawa, T., Willeke, M. and Walde, P. (2009) “Vesicles as soft templates for the enzymatic polymerization on aniline“ Langmuir, 25, 11390-11405.
Kato, K., Walde, P., Koike, N., Ichikawa, S., Ishikawa, T., Nagayama, R., Ishihara, T., Tsuji, T., Shudou, M., Omokawa, Y., Kuroiwa, T. (2008) “Temperature-sensitive nonionic vesicles prepared from Span 80 (sorbitan monooleate)” Langmuir, 24, 10762-10770.
Nishiyama, M., Ishikawa, T., Rechsteiner, H. and Glockshuber, R. (2008) “Reconstruction of pilus assembly reveals a bacterial outer membrane catalyst” Science 320, 376-379.
Capone S., Walde P., Seebach D., Ishikawa T. and Caputo R. (2008) “pH-sensitive vesicles containing a lipidic beta-amino acid with two hydrophobic chains” Chem. Biodivers. 5, 16-30.
Zepik, H.H., Walde, P., and Ishikawa, T. (2008) “Vesicle formation from reactive surfactants“ Angew. Chem. Int. Ed. Engl. 47, 1323-1325.
Namani, T., Ishikawa, T., Morigaki, K. and Walde, P. (2007) “Vesicles from docosahexaenoic acid“ Colloids Surf. B Biointerfaces 54, 118-123.
Schaffitzel, C., Oswald, M., Berger, I., Ishikawa, T., Abrahams, J.P., Koerten, H.K., Koning, R.I. and Ban, N. (2006) “Structure of the E. coli signal recognition particle bound to a translating ribosome” Nature 444, 503-506.
List of Publications
2011 - present
-
Yamamoto R, Sahashi Y, Shimo-Kon R, Sakato-Antoku M, Suzuki M, Luo L, et al.
Chlamydomonas FBB18 is a ubiquitin-like protein essential for the cytoplasmic preassembly of various ciliary dyneins
Proceedings of the National Academy of Sciences of the United States of America PNAS. 2025; 122(12): e2423948122 (10 pp.). https://doi.org/10.1073/pnas.2423948122
DORA PSI -
de Ceuninck van Capelle C, Luo L, Leitner A, Tschanz SA, Latzin P, Ott S, et al.
Proteomic and structural comparison between cilia from primary ciliary dyskinesia patients with a DNAH5 defect
Frontiers in Molecular Biosciences. 2025; 12: 1593810 (16 pp.). https://doi.org/10.3389/fmolb.2025.1593810
DORA PSI -
Alvarez N, Bruggmann R, Buchmann N, Dessimoz C, Faso C, Hofmann S, et al.
Biology community roadmap 2024. Update of Swiss community needs for research infrastructures 2029-2032
Bern: Swiss Academy of Sciences (SCNAT); 2024. Swiss Academies reports: 19/6. https://doi.org/10.5281/zenodo.14264965
DORA PSI -
Zimmermann N, Ishikawa T
Comparative structural study on axonemal and cytoplasmic dyneins
Cytoskeleton. 2024; 81(11): 681-690. https://doi.org/10.1002/cm.21897
DORA PSI -
Ishikawa T
Architecture of intraflagellar transport complexes
Nature Structural and Molecular Biology. 2023; 30(5): 570-573. https://doi.org/10.1038/s41594-023-00986-w
DORA PSI -
Ishikawa T
Cryo-electron tomography
In: Bradshaw RA, Hart GW, Stahl PD, eds. Organizational aspects of cell biology - part 1. Encyclopedia of cell biology. Amsterdam: Elsevier; 2023:28-36. https://doi.org/10.1016/B978-0-12-821618-7.00084-5
DORA PSI -
Ishikawa T
Mass-spec, cryo-EM and AI join forces for a close look at the transporter complex in cilia
EMBO Journal. 2023; 42: e113010 (3 pp.). https://doi.org/10.15252/embj.2022113010
DORA PSI -
Yildiz A, Ishikawa T
Dyneins
In: Bradshaw RA, Hart GW, Stahl PD, eds. Organizational aspects of cell biology - part 2. Encyclopedia of cell biology. Amsterdam: Elsevier; 2023:110-137. https://doi.org/10.1016/B978-0-12-821618-7.00094-8
DORA PSI -
Cvjetan N, Schuler LD, Ishikawa T, Walde P
Optimization and enhancement of the peroxidase-like activity of hemin in aqueous solutions of sodium dodecylsulfate
ACS Omega. 2023; 8(45): 42878-42899. https://doi.org/10.1021/acsomega.3c05915
DORA PSI -
Zimmermann N, Noga A, Obbineni JM, Ishikawa T
ATP-induced conformational change of axonemal outer dynein arms revealed by cryo-electron tomography
EMBO Journal. 2023; 42(12): e112466 (15 pp.). https://doi.org/10.15252/embj.2022112466
DORA PSI -
Noga A, Horii M, Goto Y, Toyooka K, Ishikawa T, Hirono M
Bld10p/Cep135 determines the number of triplets in the centriole independently of the cartwheel
EMBO Journal. 2022; 41(20): e104582 (14 pp.). https://doi.org/10.15252/embj.2020104582
DORA PSI -
Ishikawa T
Structure of motile cilia
In: Harris RJ, Marles-Wright J, eds. Macromolecular protein complexes IV. Structure and function. Subcellular biochemistry. Cham: Springer Nature; 2022:471-494. https://doi.org/10.1007/978-3-031-00793-4_15
DORA PSI -
Yamamoto R, Hwang J, Ishikawa T, Kon T, Sale WS
Composition and function of ciliary inner-dynein-arm subunits studied in Chlamydomonas reinhardtii
Cytoskeleton. 2021; 78(3): 77-96. https://doi.org/10.1002/cm.21662
DORA PSI -
Panneels V, Diaz A, Imsand C, Guizar-Sicairos M, Müller E, Bittermann AG, et al.
Imaging of retina cellular and subcellular structures using ptychographic hard X-ray tomography
Journal of Cell Science. 2021; 134(19): jcs258561 (8 pp.). https://doi.org/10.1242/jcs.258561
DORA PSI -
Tran HT, Lucas MS, Ishikawa T, Shahmoradian SH, Padeste C
A compartmentalized neuronal cell-culture platform compatible with cryo-fixation by high-pressure freezing for ultrastructural imaging
Frontiers in Neuroscience. 2021; 15: 726763 (15 pp.). https://doi.org/10.3389/fnins.2021.726763
DORA PSI -
Kutomi O, Yamamoto R, Hirose K, Mizuno K, Nakagiri Y, Imai H, et al.
A dynein-associated photoreceptor protein prevents ciliary acclimation to blue light
Science Advances. 2021; 7(9): eabf3621 (12 pp.). https://doi.org/10.1126/sciadv.abf3621
DORA PSI -
Poghosyan E, Iacovache I, Faltova L, Leitner A, Yang P, Diener DR, et al.
The structure and symmetry of the radial spoke protein complex in Chlamydomonas flagella
Journal of Cell Science. 2020; 133(16): jcs245233 (9 pp.). https://doi.org/10.1242/jcs.245233
DORA PSI -
Rösner B, Finizio S, Koch F, Döring F, Guzenko VA, Langer M, et al.
Soft x-ray microscopy with 7 nm resolution
Optica. 2020; 7(11): 1602-1608. https://doi.org/10.1364/OPTICA.399885
DORA PSI -
Li T, Krumeich F, Ihli J, Ma Z, Ishikawa T, Pinar AB, et al.
Heavy atom labeling enables silanol defect visualization in silicalite-1 crystals
Chemical Communications. 2019; 55(4): 482-485. https://doi.org/10.1039/c8cc07912a
DORA PSI -
Kashima K, Fujisaki T, Serrano-Luginbühl S, Kissner R, Janošević Ležaić A, Bajuk-Bogdanović D, et al.
Effect of template type on the Trametes versicolor laccase-catalyzed oligomerization of the aniline dimer p-aminodiphenylamine (PADPA)
ACS Omega. 2019; 4(2): 2931-2947. https://doi.org/10.1021/acsomega.8b03441
DORA PSI -
Zhu X, Poghosyan E, Rezabkova L, Mehall B, Sakakibara H, Hirono M, et al.
The roles of a flagellar HSP40 ensuring rhythmic beating
Molecular Biology of the Cell. 2019; 30(2): 228-241. https://doi.org/10.1091/mbc.E18-01-0047
DORA PSI -
Guerrero-Ferreira RC, Hupfeld M, Nazarov S, Taylor NMI, Shneider MM, Obbineni JM, et al.
Structure and transformation of bacteriophage A511 baseplate and tail upon infection of Listeria cells
EMBO Journal. 2019; 38(3): e99455 (20 pp.). https://doi.org/10.15252/embj.201899455
DORA PSI -
Neuhaus F, Mueller D, Tanasescu R, Balog S, Ishikawa T, Brezesinski G, et al.
Synthesis and biophysical characterization of an odd-numbered 1,3-diamidophospholipid
Langmuir. 2018; 34(10): 3215-3220. https://doi.org/10.1021/acs.langmuir.7b04227
DORA PSI -
Isabettini S, Stucki S, Massabni S, Baumgartner ME, Reckey PQ, Kohlbrecher J, et al.
Development of smart optical gels with highly magnetically responsive bicelles
ACS Applied Materials and Interfaces. 2018; 10(10): 8926-8936. https://doi.org/10.1021/acsami.7b17134
DORA PSI -
Buscema M, Matviykiv S, Gerganova G, Mészáros T, Kozma GT, Mettal U, et al.
Immunocompatibility of Rad-PC-Rad liposomes in vitro, based on human complement activation and cytokine release
Precision Nanomedicine. 2018; 1(1): 43-62. https://doi.org/10.29016/180419.2
DORA PSI -
Neuhaus F, Mueller D, Tanasescu R, Stefaniu C, Zaffalon P-L, Balog S, et al.
Against the rules: pressure induced transition from high to reduced order
Soft Matter. 2018; 14(19): 3978-3986. https://doi.org/10.1039/c8sm00212f
DORA PSI -
Ishikawa T
Organization of dyneins in the axoneme
In: King SM, ed. Dyneins: The biology of dynein motors. Amsterdam: Elsevier; 2018:203-217. https://doi.org/10.1016/B978-0-12-809471-6.00006-1
DORA PSI -
Isabettini S, Baumgartner ME, Reckey PQ, Kohlbrecher J, Ishikawa T, Fischer P, et al.
Methods for generating highly magnetically responsive lanthanide-chelating phospholipid polymolecular assemblies
Langmuir. 2017; 33(25): 6363-6371. https://doi.org/10.1021/acs.langmuir.7b00725
DORA PSI -
Yamamoto R, Obbineni JM, Alford LM, Ide T, Owa M, Hwang J, et al.
Chlamydomonas DYX1C1/PF23 is essential for axonemal assembly and proper morphology of inner dynein arms
PLoS Genetics. 2017; 13(10): e1006996 (21 pp.). https://doi.org/10.1371/journal.pgen.1006996
DORA PSI -
Isabettini S, Massabni S, Hodzic A, Durovic D, Kohlbrecher J, Ishikawa T, et al.
Molecular engineering of lanthanide ion chelating phospholipids generating assemblies with a switched magnetic susceptibility
Physical Chemistry Chemical Physics. 2017; 19(31): 20991-21002. https://doi.org/10.1039/c7cp03994h
DORA PSI -
Zhu X, Poghosyan E, Gopal R, Liu Y, Ciruelas KS, Maizy Y, et al.
General and specific promotion of flagellar assembly by a flagellar nucleoside diphosphate kinase
Molecular Biology of the Cell. 2017; 28(22): 3029-3042. https://doi.org/10.1091/mbc.E17-03-0156
DORA PSI -
Obbineni JM, Yamamoto R, Ishikawa T
A simple and fast approach for missing-wedge invariant classification of subtomograms extracted from filamentous structures
Journal of Structural Biology. 2017; 197(2): 145-154. https://doi.org/10.1016/j.jsb.2016.08.003
DORA PSI -
Buscema M, Matviykiv S, Mészáros T, Gerganova G, Weinberger A, Mettal U, et al.
Immunological response to nitroglycerin-loaded shear-responsive liposomes in vitro and in vivo
Journal of Controlled Release. 2017; 264: 14-23. https://doi.org/10.1016/j.jconrel.2017.08.010
DORA PSI -
Neuhaus F, Mueller D, Tanasescu R, Balog S, Ishikawa T, Brezesinski G, et al.
Vesicle origami: cuboid phospholipid vesicles formed by template-free self-assembly
Angewandte Chemie International Edition. 2017; 56(23): 6515-6518. https://doi.org/10.1002/anie.201701634
DORA PSI -
Isabettini S, Liebi M, Kohlbrecher J, Ishikawa T, Fischer P, Windhab EJ, et al.
Mastering the magnetic susceptibility of magnetically responsive bicelles with 3β-amino-5-cholestene and complexed lanthanide ions
Physical Chemistry Chemical Physics. 2017; 19(17): 10820-10824. https://doi.org/10.1039/c7cp01025g
DORA PSI -
Ishikawa T
Axoneme structure from motile cilia
Cold Spring Harbor Perspectives in Biology. 2017; 9(1): a028076 (18 pp.). https://doi.org/10.1101/cshperspect.a028076
DORA PSI -
Cypranowska CA, Yildiz A, Ishikawa T
Dyneins
In: Bradshaw RA, Stahl PD, eds. Encyclopedia of cell biology. Reference module in biomedical sciences. Amsterdam: Elsevier; 2016:620-636. https://doi.org/10.1016/B978-0-12-394447-4.20101-6
DORA PSI -
Isabettini S, Liebi M, Kohlbrecher J, Ishikawa T, Windhab EJ, Fischer P, et al.
Tailoring bicelle morphology and thermal stability with lanthanide-chelating cholesterol conjugates
Langmuir. 2016; 32(35): 9005-9014. https://doi.org/10.1021/acs.langmuir.6b01968
DORA PSI -
Ishikawa T
Electron tomography
In: Bradshaw RA, Stahl PD, eds. Encyclopedia of cell biology. Reference module in biomedical sciences. Amsterdam: Elsevier; 2016:22-31. https://doi.org/10.1016/B978-0-12-394447-4.20006-0
DORA PSI -
Tanasescu R, Lanz MA, Mueller D, Tassler S, Ishikawa T, Reiter R, et al.
Vesicle origami and the influence of cholesterol on lipid packing
Langmuir. 2016; 32(19): 4896-4903. https://doi.org/10.1021/acs.langmuir.6b01143
DORA PSI -
Aroua S, Tiu EGV, Ishikawa T, Yamakoshi Y
Well-defined amphiphilic C60-PEG conjugates: water-soluble and thermoresponsive materials
Helvetica Chimica Acta. 2016; 99(10): 805-813. https://doi.org/10.1002/hlca.201600171
DORA PSI -
Liebi M, Kuster S, Kohlbrecher J, Ishikawa T, Walde P, Windhab EJ, et al.
Design of magnetically responsive phospholipid bicelles towards switchable optical hydrogels
Swiss Neutron News. https://sgn.web.psi.ch/sgn/snn.html. Published 2015. Accessed no date.
DORA PSI -
Diaz A, Malkova B, Holler M, Guizar-Sicairos M, Lima E, Panneels V, et al.
Three-dimensional mass density mapping of cellular ultrastructure by ptychographic X-ray nanotomography
Journal of Structural Biology. 2015; 192(3): 461-469. https://doi.org/10.1016/j.jsb.2015.10.008
DORA PSI -
Maheshwari A, Obbineni JM, Bui KH, Shibata K, Toyoshima YY, Ishikawa T
α- and β-Tubulin Lattice of the Axonemal Microtubule Doublet and Binding Proteins Revealed by Single Particle Cryo-Electron Microscopy and Tomography
Structure. 2015; 23(9): 1584-1595. https://doi.org/10.1016/j.str.2015.06.017
DORA PSI -
Fodor D, Ishikawa T, Krumeich F, van Bokhoven JA
Synthesis of single crystal nanoreactor materials with multiple catalytic functions by incipient wetness impregnation and ion exchange
Advanced Materials. 2015; 27(11): 1919-1923. https://doi.org/10.1002/adma.201404628
DORA PSI -
Aroua S, Tiu EGV, Ayer M, Ishikawa T, Yamakoshi Y
RAFT synthesis of poly(vinylpyrrolidone) amine and preparation of a water-soluble C60-PVP conjugate
Polymer Chemistry. 2015; 6(14): 2616-2619. https://doi.org/10.1039/c4py01333f
DORA PSI -
Weinberger A, Tanasescu R, Stefaniu C, Fedotenko LA, Favarger F, Ishikawa T, et al.
Bilayer properties of 1,3-diamidophospholipids
Langmuir. 2015; 31(6): 1879-1884. https://doi.org/10.1021/la5041745
DORA PSI -
Ishikawa T
Cryo-electron tomography of motile cilia and flagella
Cilia. 2015; 4(Suppl. 1): 3 (20 pp.). https://doi.org/10.1186/s13630-014-0012-7
DORA PSI -
Liebi M, Kuster S, Kohlbrecher J, Ishikawa T, Fischer P, Walde P, et al.
Magnetically enhanced bicelles delivering switchable anisotropy in optical gels
ACS Applied Materials and Interfaces. 2014; 6(2): 1100-1105. https://doi.org/10.1021/am4046469
DORA PSI -
Ueno H, Bui KH, Ishikawa T, Imai Y, Yamaguchi T, Ishikawa T
Structure of dimeric axonemal dynein in cilia suggests an alternative mechanism of force generation
Cytoskeleton. 2014; 71(7): 412-422. https://doi.org/10.1002/cm.21180
DORA PSI -
Ishikawa T
Protein tagging reveals new insights into signaling in flagella
Journal of Cell Biology. 2014; 204(5): 631-633. https://doi.org/10.1083/jcb.201401142
DORA PSI -
Ishikawa T
3D structure of eukaryotic flagella/cilia by cryo-electron tomography
Biophysics. 2013; 9: 141-148. https://doi.org/10.2142/biophysics.9.141
DORA PSI -
Bui KH, Ishikawa T
3D structural analysis of flagella/cilia by cryo-electron tomography
In: Marshall WF, ed. Cilia, part A. Methods in enzymology. Amsterdam: Elsevier; 2013:305-323. https://doi.org/10.1016/B978-0-12-397945-2.00017-2
DORA PSI -
Liebi M, Kuster S, Kohlbrecher J, Ishikawa T, Fischer P, Walde P, et al.
Cholesterol-diethylenetriaminepentaacetate complexed with thulium ions integrated into bicelles to increase their magnetic alignability
Journal of Physical Chemistry B. 2013; 117(47): 14743-14748. https://doi.org/10.1021/jp406599c
DORA PSI -
Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T
Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme
Journal of Cell Biology. 2012; 198(5): 913-925. https://doi.org/10.1083/jcb.201201120
DORA PSI -
Liebi M, Kohlbrecher J, Ishikawa T, Fischer P, Walde P, Windhab EJ
Cholesterol increases the magnetic aligning of bicellar disks from an aqueous mixture of DMPC and DMPE-DTPA with complexed thulium ions
Langmuir. 2012; 28(29): 10905-10915. https://doi.org/10.1021/la3019327
DORA PSI -
Maheshwari A, Ishikawa T
Heterogeneity of dynein structure implies coordinated suppression of dynein motor activity in the axoneme
Journal of Structural Biology. 2012; 179(2): 235-241. https://doi.org/10.1016/j.jsb.2012.04.018
DORA PSI -
Ishikawa T
Structural biology of cytoplasmic and axonemal dyneins
Journal of Structural Biology. 2012; 179(2): 229-234. https://doi.org/10.1016/j.jsb.2012.05.016
DORA PSI -
Lowell AN, Qiao H, Liu T, Ishikawa T, Zhang H, Oriana S, et al.
Functionalized low-density lipoprotein nanoparticles for in vivo enhancement of atherosclerosis on magnetic resonance images
Bioconjugate Chemistry. 2012; 23(11): 2313-2319. https://doi.org/10.1021/bc300561e
DORA PSI -
Tokutsu R, Kato N, Bui KH, Ishikawa T, Minagawa J
Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii
Journal of Biological Chemistry. 2012; 287(37): 31574-31581. https://doi.org/10.1074/jbc.M111.331991
DORA PSI -
Ueno H, Ishikawa T, Bui KH, Gonda K, Ishikawa T, Yamaguchi T
Mouse respiratory cilia with the asymmetric axonemal structure on sparsely distributed ciliary cells can generate overall directional flow
Nanomedicine: Nanotechnology, Biology and Medicine. 2012; 8(7): 1081-1087. https://doi.org/10.1016/j.nano.2012.01.004
DORA PSI -
Pigino G, Maheshwari A, Bui KH, Shingyoji C, Kamimura S, Ishikawa T
Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins
Journal of Structural Biology. 2012; 178(2): 199-206. https://doi.org/10.1016/j.jsb.2012.02.012
DORA PSI -
Ueno H, Ishikawa T, Bui KH, Gonda K, Ishikawa T, Yamaguchi T
Analysis of ciliary motion and the axonemal structure in the mouse respiratory cilia
In: ASME 2012 summer bioengineering conference, SBC 2012. ; 2012:(2 pp.). https://doi.org/10.1115/SBC2012-80232
DORA PSI -
Junker K, Zandomeneghi G, Guo Z, Kissner R, Ishikawa T, Kohlbrecher J, et al.
Mechanistic aspects of the horseradish peroxidase-catalysed polymerisation of aniline in the presence of AOT vesicles as templates
RSC Advances. 2012; 2(16): 6478-6495. https://doi.org/10.1039/c2ra20566a
DORA PSI -
Guo Z, Hauser N, Moreno A, Ishikawa T, Walde P
AOT vesicles as templates for the horseradish peroxidase-triggered polymerization of aniline
Soft Matter. 2011; 7(1): 180-193. https://doi.org/10.1039/c0sm00599a
DORA PSI -
Bui KH, Pigino G, Ishikawa T
Three-dimensional structural analysis of eukaryotic flagella/cilia by electron cryo-tomography
Journal of Synchrotron Radiation. 2011; 18(1): 2-5. https://doi.org/10.1107/S0909049510036812
DORA PSI -
Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T
Cryoelectron tomography of radial spokes in cilia and flagella
Journal of Cell Biology. 2011; 195(4): 673-687. https://doi.org/10.1083/jcb.201106125
DORA PSI -
Megli FM, Conte E, Ishikawa T
Cholesterol attenuates and prevents bilayer damage and breakdown in lipoperoxidized model membranes. A spin labeling EPR study
Biochimica et Biophysica Acta: Biomembranes. 2011; 1808(9): 2267-2274. https://doi.org/10.1016/j.bbamem.2011.04.016
DORA PSI -
Ishikawa T
Organization of dyneins in the axoneme
In: King SM, ed. Dyneins. Structure, biology and disease. Elsevier; 2011:244-271. https://doi.org/10.1016/B978-0-12-382004-4.10008-1
DORA PSI -
Ishikawa T
3D structures of axonemes
In: Hirose K, ed. Handbook of Dynein. New York: Jenny Stanford Publishing; 2011:245-266. https://doi.org/10.1201/b11622
DORA PSI
2006-2010
-
Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis
NATURE STRUCTURAL & MOLECULAR BIOLOGY 17, 761 (2010).DOI: 10.1038/nsmb.1832
-
Three-dimensional structural analysis of eukaryotic flagella/cilia by electron cryo-tomography
JOURNAL OF SYNCHROTRON RADIATION 18, 2 (2010).DOI: 10.1107/s0909049510036812
- Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella
The Journal of Cell Biology 186, 437 (2009).DOI: 10.1083/jcb.200903082
-
Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella
The Journal of Cell Biology 183, 923 (2008).DOI: 10.1083/jcb.200808050
- Structure of the E. coli signal recognition particle bound to a translating ribosome
NATURE 448, 1076 (2007).DOI: 10.1038/nature06169
- Structure of the E. coli signal recognition particle bound to a translating ribosome
NATURE 444, 503 (2006).DOI: 10.1038/nature05182
before 2005
Ishikawa, T., Maurizi, M.R. and Steven, A.C. (2004) “The N-terminal substrate-binding domain of ClpA unfoldase is highly mobile and extends axially from the distal surface of ClpAP protease” J. Struct. Biol. 146, 180-188.
Zakalskiy, A., Hoegenauer, G., Ishikawa, T., Wehrschuetz-Sigl, E., Wendler, F., Teis, D. Zisser, D., Steven, A.C. and Bergler, H. (2002) “Structural and enzymatic properties of the AAA Protein Drg1p from Saccharomyces cerevisiae. Decoupling of intracellular function from ATPase activity and hexamerization” J. Biol. Chem. 277, 26788-26795.
Miyanishi, T., Ishikawa, T., Hayashibara, T., Maita, T. and Wakabayashi, T. (2002) “The two actin-binding regions on the myosin heads of cardiac muscle” Biochemistry 41, 5429-5438.
Ishikawa, T., Beuron, F., Kessel, M., Wickner, S., Maurizi, M.R. and Steven, A.C. (2001) “Translocation pathway of protein substrates in ClpAP protease” Proc. Natl. Acad. Sci. USA 98, 4328-33.
Narita, A., Yasunaga, T., Ishikawa, T., Mayanagi, K. and Wakabayashi, T. (2001) “Ca(2+)-induced switching of troponin and tropomyosin on actin filaments as revealed by electron cryo-microscopy” J. Mol. Biol. 308, 241-261.
Singh, S.K., Rozycki, J., Ortega, J., Ishikawa, T., Lo, J., Steven, A.C. and Maurizi, M.R. (2001) “Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis” J. Biol. Chem. 276, 29420-29429.
Ishikawa, T., Maurizi, M.R., Belnap, D.M. and Steven, A.C. (2000) “Docking of components in a bacterial complex” Nature 408, 667-668.
Ortega, J., Singh, S.K., Ishikawa, T., Maurizi, M.R. and Steven, A.C. (2000) “Visualization of substrate binding and translocation by the ATP-dependent protease,ClpXP” Mol. Cell 6, 1515-1521.
Ishikawa, T. and Wakabayashi, T. (1999) “Calcium-induced changes in the location and conformation of troponin in skeletal muscle thin filaments” J. Biochem. (Tokyo) 126, 200-211.
Mayanagi, K., Ishikawa, T., Toyoshima, C., Inoue, Y. and Nakazato, K. (1998) “Three- Dimensional Electron Microscopy of the Photosystem II Core Complex” J. Struct. Biol. 123, 211-224.
Kikkawa, M., Ishikawa, T., Wakabayashi, T. and Hirokawa, N. (1995) “Threedimensional structure of the kinesin head-microtubule complex” Nature 376, 274-277.
Ishikawa, T. and Wakabayashi, T. (1995) “Proposal of alignment-independent classification of electron microscopic images with helical symmetry and its application to reconstituted thin filament of skeletal muscle” Ultramicroscopy 57, 91-101.
Ishikawa, T. and Wakabayashi, T. (1994) “Calcium induced change in three-dimesional structure of thin filaments of rabbit skeletal muscle as revealed by cryo-elecron miscroscopy” Biochem. Biophys. Res. Commun. 203, 951-958.
Kikkawa, M., Ishikawa, T., Nakata, T., Wakabayashi, T. and Hirokawa, N. (1994) “Direct visualization of the microtubule lattice seam both in vitro and in vivo” J. Cell Biol. 127, 1965-1971.