Peering into proteins for better drug discovery
(Graphic: Thomas Splettstösser)
The triggering mechanism of a critical chemokine receptor has finally been caught on camera.
Researchers have used molecular imaging to reveal the activation mechanism of CCR5, a crucial chemokine receptor found on white blood cells and T cells. CCR5 is involved in immune-related processes around cancer and neuroinflammation, and is one of the cellular entry points for HIV. “Our team has a long-standing interest in this receptor because it is such a significant drug target,” says Xavier Deupi from the Condensed Matter Theory Group at the Paul Scherrer Institute (PSI). “And yet, its activation mechanism is poorly understood.”
Cell-surface receptors like CCR5 detect signalling proteins, called chemokines, then induce cell responses. Chemokines bind to the receptor’s extracellular domains and insert their amino (N) terminus to deliver the activation signal. The receptor then releases a G-protein trimer — three large molecules that act like a switch to transmit or block cellular signalling pathways.
Two chemokine receptors have already been observed bound to a G protein. They displayed a shallow chemokine binding, meaning activation must be triggered from the outer edges of the receptor. CCR5, however, is thought to respond to chemokines with longer N termini, necessitating a different activation mechanism. The transient and unstable nature of receptor/G protein complexes has made researching this theory tricky.
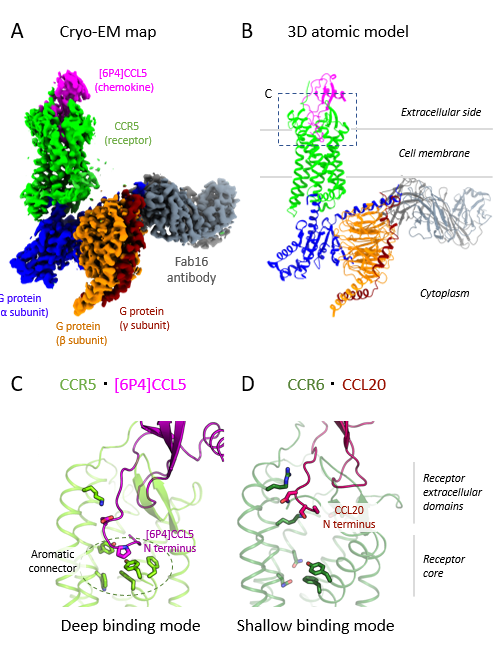
Researchers from PSI and several other Swiss institutions solved this problem by mixing human CCR5 with a highly potent variant of its natural chemokine ([6P4]CCL5), a Gi heterotrimer, and an antibody. “A crucial prerequisite for cryo-electron microscopy (cryo-EM) is a complex that can endure the harsh process of sample preparation,” explains Deupi. “So, although this antibody isn’t part of CCR5/G protein complexes in human cells, it was key to achieving the stability required for our experiments.”
Cold hard truths
Using cryo-EM, which freezes the sample below –180°C, the researchers were able to capture thousands of images of their complex from different angles. They converted the images into a 3D map, enabling them to solve and analyse the molecular structure. The map revealed that features in certain regions of the complex were less defined, suggesting that these molecular components had more flexibility to move when activated.
The researchers identified two key sites where [6P4]CCL5 and CCR5 interact: one connecting the chemokine core to the receptor’s outer edge, which controls insertion depth, and one at the receptor’s binding pocket. They observed that the [6P4]CCL5 N terminus extends deeper into CCR5’s binding pocket than for other chemokine–receptor pairs, enabling unique interactions to trigger activation (see figure).
Next, the team plans to solve the structure of CCR5 bound to wild-type CCL5, the natural chemokine, instead of the potent experimental variant. “We speculate that wild-type CCL5 has a similar binding pose,” says Deupi. “A new structural study would provide useful additional data to better understand the activation mechanism of chemokine receptors.”
By uncovering new and unique binding and activation modes, the team hopes to rationalize the relationship between sequence, structure and activity of chemokines and their receptors. “Our findings have shed new light on the molecular pharmacology of chemokine receptors,” says Deupi, “which will aid drug discovery for these important pharmaceutical targets.”