The macromolecular crystallography (MX) group at the Swiss Light Source operates three beamlines (PXI, PXII and PXIII), which allow study of biological molecules such as proteins, viruses and nucleic acids (RNA and DNA). In the macromolecular crystallography experiment high intensity incident X-rays are diffracted on a crystalline sample giving rise to a regular pattern of sharp spots. Analysis of the intensities and positions of the diffraction spots allow determination of high-resolution three-dimensional arrangement of atoms in space (macromolecular structure). MX technique enables research, which aims at 1) understanding function and mechanism of macromolecules in cells and organisms; 2) structure-based drug design utilizing high-throughput screening (HTC) and/or fragment-based drug design (FBDD) approaches.
To aid academic and industrial drug design projects utilizing FBDD with X-ray crystallography (xFBDD) as primary technique, the SLS MX group has recently established the fast fragment and compound-screening (FFCS) pipeline. The goal of the FFCS project was to create an integrated next generation pipeline for high throughput crystal soaking, handling and data collection, which allows crystallography-based screening of protein crystals against hundreds of potential drug leads. The project created tools that make the process faster, more efficient, more automated and thereby more appealing to users. The resulting FFCS pipeline has been successfully applied to support analysis of small-molecule ligands of the human YTHDC1 domain that recognizes N6-methylated adenine (m6A) in RNA (1).
Coronaviruses are RNA viruses, which cause significant percentage of acute respiratory illnesses in humans (SARS, MERS & COVID-19 pandemics of deadly pneumonia). No vaccines or specific antiviral treatment are currently available against Coronaviruses. Structural characterization of proteins essential in virus life cycle provides valid functional information and is the basis for structure-based drug design.
The SLS MX group at the Photon Science Division at PSI collaborates with various Chinese groups on the X-ray structure determination of viral proteins and their complexes with inhibitors. Currently research on coronaviruses is still relatively limited, despite three pandemic outbreaks within the last 15 years, i.e. 2003 SARS, 2012/2015 MERS, 2019/20 COVID-19. The development of the antiviral treatment is in (obvious) demand and presents a unique research opportunity. The MX beamlines and crystallization facility at PSI will provide priority access to COVID-19 related structures-based drug discovery projects.
First MX results of the priority COVID-19 call
Beamline X06SA-PXI
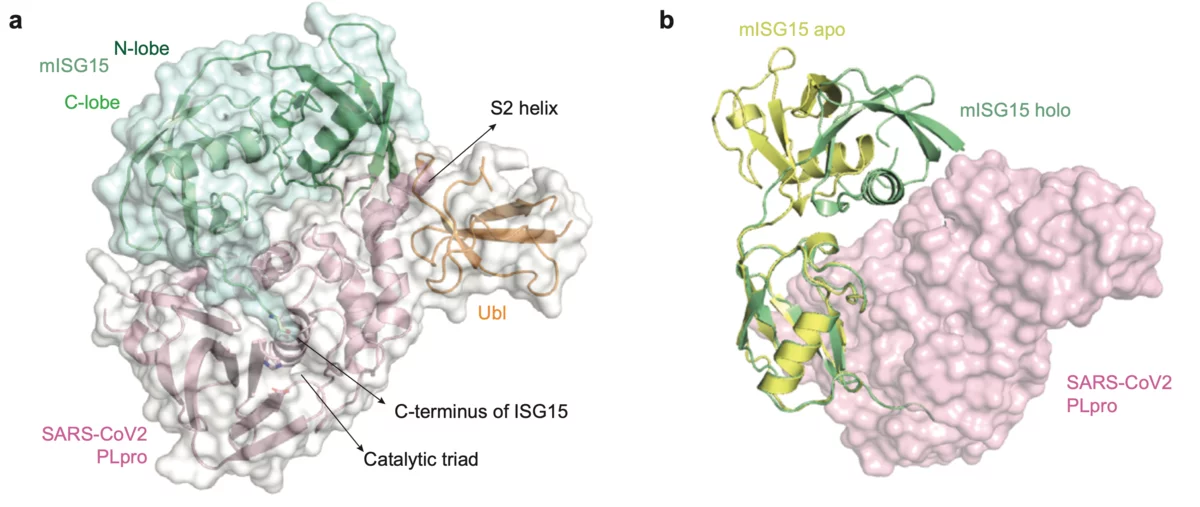
Scientists from the Goethe University in Frankfurt am Main, Germany have published results on the papain-like protease (PLpro), an essential enzyme of SARS-CoV-2. The structural biology work was performed at the macromolecular crystallography beamline X06SA-PXI at SLS following the opening of the "PRIORITY COVID-19 Call”. The paper was submitted within one month after answering the proposal call. The crystallographic data collection happened on the 9th of April after the planned Easter shutdown of the SLS was cancelled for this specific experiment.
For the full article, please click here
Two protein structures from coronavirus determined at PX beamline at SLS
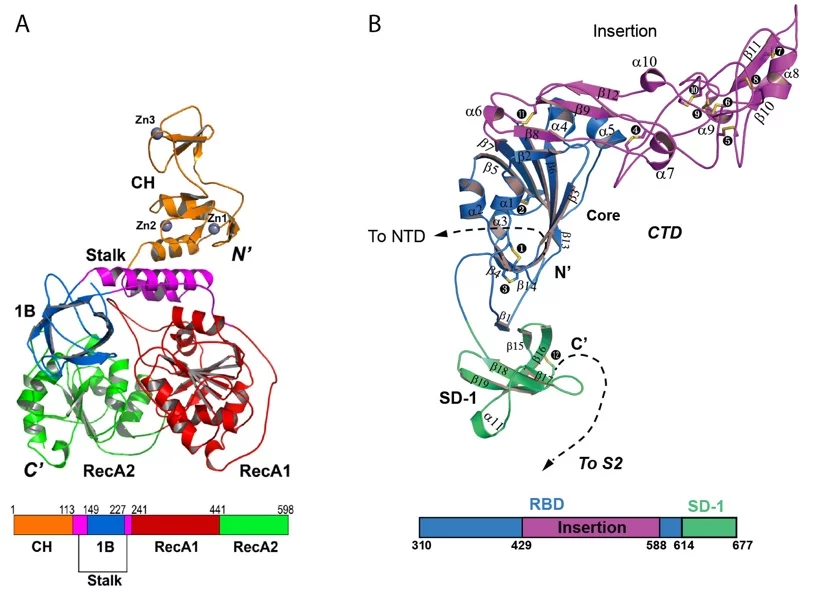
The SLS MX group participated in the structural characterization of the MERS coronavirus nsp13 helicase, which is one of 3 evolutionary most conserved proteins in nidoviruses and a central component of virus replication-transcription complex ((2); Figure 1A). Moreover, structure of the human betacoronavirus HKU1 protein S1, which docks virus to the cells of infected person, was solved with experimental phasing technique called native-SAD using data collected at the SLS MX beamline X06DA ((3); Figure 1B)
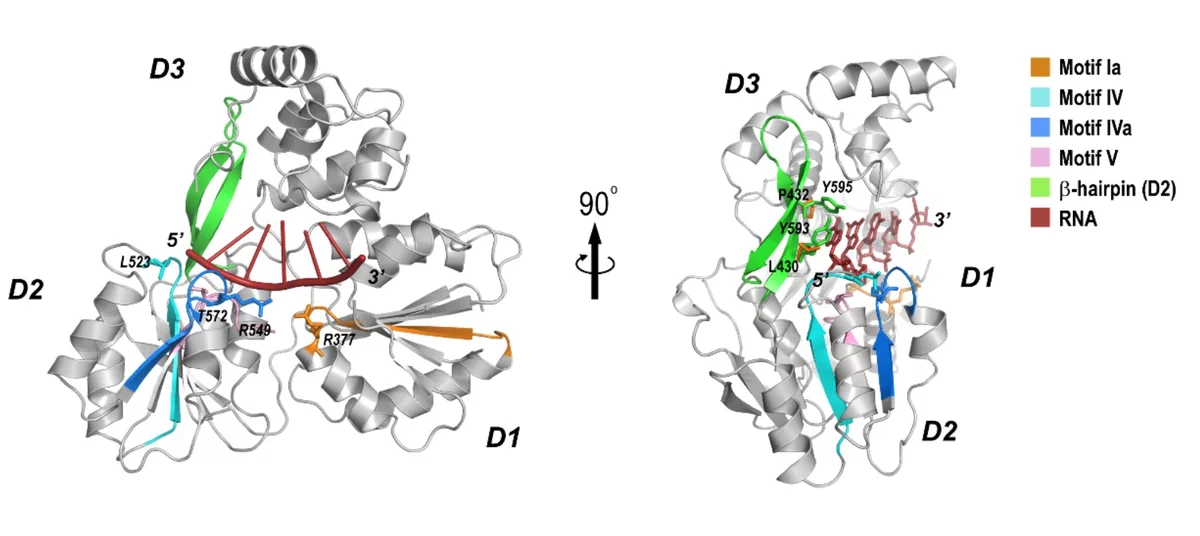
Crystal structure of the NS3-like helicase from Alongshan virus
THe Alongshan virus (ALSV) is an emerging human pathogen recently identified in China and rapidly spreading to the European continent in 2019 raising concerns about public health. ALSV belongs to the distinct Jingmenvirus group within the Flaviviridae family. The X-ray structure, generated by the MX group at PSI, provides a structural framework for drug design and suggest a possibility of developing broad-spectrum antiviral drug against both Flavivirus and Jingmenvirus.
Xiaopan Liu, Kaixiang Zhu, Justyna Aleksandra Wojdyla, Pu Chen, Bo Qin, Ziheng Li, Meitian Wang, and Sheng Cui, IUCr Journal, under revision
(1) Bedi RK, Huang D, Wiedmer L, Li Y, Dolbois A, Wojdyla JA, Sharpe ME, Caflisch A, Sledz P, 2020, Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments, ACS Chem Biol 15, 3, 618-625, https://doi.org/10.1021/acschembio.9b00894
(2) Hao W, Wojdyla JA, Zhao R, Han R, Das R, Zlatev I, Manoharan M, Wang M, Cui S, 2017, Crystal structure of Middle Eastrespiratory syndrome coronavirus helicase, PLoS Pathogens 26;13(6):e1006474, https://doi.org/10.1371/journal.ppat.1006474
(3) Ou X, Guan H, Qin B, Mu Z, Wojdyla J, Wang M, Dominguez SR, Qian Z, Cui S, 2017, Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1, Nature Communications 8, 15216, https://doi.org/10.1038/ncomms15216
Contact
Photon Science Division
Paul Scherrer Institut
5232 Villigen-PSI
Switzerland
Head of MX and Group Leader (i.p.) MX Applications
Laboratory for Macromolecules and Bioimaging LSB
Dr Meitian Wang